mjpdejong
Shared posts
Monsanto Cultivates a Rose That Doesn’t Wilt
The Quiet Crisis Unfolding in Software Development
|
Sjon
shared this story
from |
Comments URL: https://news.ycombinator.com/item?id=11791444
Automating DNA origami opens door to many new uses
Why ABN Amro Wants to Separate Bitcoin from the Blockchain
Washington Grapples with a Thorny Question: What Is a GMO Anyway?
Oxford Nanopore's London Calling Kicks Off
Oxford Nanopore Introduces SmidgION at London Calling 2016
London Calling 2016, the two-day conference on all things Nanopore sequencing organized by Oxford Nanopore has started today. Here are storified tweets from Clive Brown’s ( CTO of Oxford Nanopore Technologies Ltd) plenary talk, one of the most anticipated talks at London calling.
One of the biggest highlights from the talk is SmidgION, the smallest sequencing device that can be plugged into a smartphone for sequencing and analysis. SmidgION will have 256 pores, can sequence for four hours with a smartphone,and produce 230 Mb/hour. SmidgION is expected to be available in late 2017.
Big Ideas, Big Conflicts in Plan to Synthesize a Human Genome
Are our gut bacteria the key to immortality?
De Winter, G. (2014) Aging as Disease. Medicine, Health Care and Philosophy, 18(2), 237-243. DOI: 10.1007/s11019-014-9600-y
Aging as DiseaseBiagi E, Franceschi C, Rampelli S, Severgnini M, Ostan R, Turroni S, Consolandi C, Quercia S, Scurti M, Monti D.... (2016) Gut Microbiota and Extreme Longevity. Current biology : CB. PMID: 27185560
Gut Microbiota and Extreme Longevity.Bitcoin Protocol Role Models
A couple of months ago, I was having Yet Another Argument with a Bitcoin Core contributor about the one megabyte block size limit.
I had asked: “what other successful Internet protocols impose arbitrary limits on themselves similar to the one-megabyte limit?”
His answer was “there are limits on the size of the global routing table in the Border Gateway Protocol (BGP) protocol.”
BGP is the protocol routers use to decide what to do when they get a data packet bound for an IP address like “111.12.1.1” – they send it to China (111.12.1.1 belongs to the China Mobile Communications Corporation right now).
So I went and skimmed through the BGP protocol documents. Then read the wikipedia page on BGP. And had the opportunity to talk with Justin Newton, who participated in the debates on Internet scalability fifteen years ago.
And found that there are no protocol-level limits on the size of BGP routing tables. There is no place in any BGP specification I can find that says “Routing Tables Shall Be No More Than Eleven Gigabytes Big.”
BGP is interesting, because it is a completely different way of doing decentralized consensus than Bitcoin. Instead of proof-of-work, BGP’s consensus is built on real-world trust relationships between people operating the networks that make up the Internet. That works surprisingly well most of the time, especially considering the stunning lack of security in the BGP protocol.
There are limits on routing table sizes, but they are not top-down-specified-in-a-standards-document protocol limits. They are organic limits that arise from whatever hardware is available and from the (sometimes very contentious!) interaction of the engineers keeping the Internet backbone up and running.
I haven’t been able to find a widely-used Internet-scale protocol that arbitrarily limits itself. The closest I’ve found is the Simple Mail Transport Protocol (SMTP) which has a SIZE header that a server can use to specify the maximum email message size it will accept. The arbitrary limit chosen by the SMTP designers is 99,999,999,999,999,999,999 bytes (just under 100 exabytes).
The HTTP 2.0 spec explicitly discusses denial-of-service attacks, but doesn’t impose hard limits: “An endpoint MAY treat activity that is suspicious as a connection error of type ENHANCE_YOUR_CALM” (I can’t help imagining the server as a California Surfer saying “enhance your calm, dude!)
That’s a well-designed spec. Trust that smart developers will fix scaling or denial-of-service issues as they arise– and, if for some unforeseen reason it turns out they can’t, trust that there will be either an amendment to the spec or a "best practices” document to fix the problem(s).
Top scientists hold closed meeting to discuss building a human genome from scratch
Over 130 scientists, lawyers, entrepreneurs, and government officials from five continents gathered at Harvard this week for an “exploratory” meeting to discuss the topic of creating genomes from scratch — including, but not limited to, those of humans, said George Church, Harvard geneticist and co-organizer of the meeting.
The meeting was closed to the press, which drew the ire of prominent academics.
Why scientists should learn to fail, fast and often
Silicon Valley runs on failure. Its unofficial motto, after all, is “Fail fast, fail often,” and it is the region that gave birth to FailCon, where stories of entrepreneurial failure are badges of honor.
That’s more than just cute marketing. As the New Yorker’s James Surowiecki has written, “In the delusions of entrepreneurs are the seeds of technological progress.” Failures, in other words, are not only acceptable, but beneficial.
WATCH: What happened to astronaut Scott Kelly’s DNA during his year in space
“I was a dorky kid,” Christopher Mason admits. “I went to Space Camp. I’d always been thinking about maybe being an astronaut.”
But not long after Space Camp, Mason discovered something even more seductive: DNA. He decided he wanted to be a geneticist instead.
What generics? Americans spent an extra $73b for brand-name meds
Doctors may be encouraged to prescribe lower-cost generics, but a new study found that Americans spent an extra $73 billion between 2010 and 2012 on pricier brand-name drugs because physicians failed to sufficiently recommend these copycat treatments to their patients. And consumers paid nearly one-third of those additional costs through out-of-pocket payments.
Notably, the study found that the excess spending occurred when a generic version was not available for a specific brand-name drug, but a doctor could have prescribed a lower-cost, copycat for a similar brand-name medicine. For example, instead of prescribing a lower-cost generic for a brand-name cholesterol pill, a doctor might have prescribed a similar brand-name cholesterol medicine.
Gene Therapy’s First Out-and-Out Cure Is Here
How Blockchain Is Helping Genomics Research
Cpf1: CRISPR-enzyme scissors cutting both RNA and DNA
Kuwaiti Government will DNA Test Everyone
|
Sjon
shared this story
from |
There's a new law that will enforce DNA testing for everyone: citizens, expatriates, and visitors. They promise that the program "does not include genealogical implications or affects personal freedoms and privacy."
I assume that "visitors" includes tourists, so presumably the entry procedure at passport control will now include a cheek swab. And there is nothing preventing the Kuwaiti government from sharing that information with any other government.
Uncovering the genetic elements that drive regeneration
Kang, J., Hu, J., Karra, R., Dickson, A., Tornini, V., Nachtrab, G., Gemberling, M., Goldman, J., Black, B., & Poss, K. (2016) Modulation of tissue repair by regeneration enhancer elements. Nature. DOI: 10.1038/nature17644
Modulation of tissue repair by regeneration enhancer elementsWhy Microsoft needed to make Windows run Linux software
|
Sjon
shared this story
from |
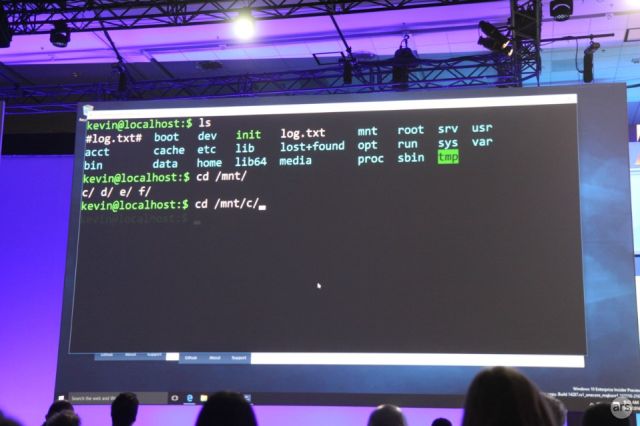
It's bash, it's Windows, it's not a virtual machine.
Perhaps the biggest surprise to come from Microsoft's Build developer conference last week was the Windows Subsystem for Linux (WSL).
The system will ship as part of this summer's Anniversary Update for Windows 10. WSL has two parts; there's the core subsystem, which is already included in Insider Preview builds of the operating system, and then a package of software that Canonical will provide. The core subsystem is what provides the Linux API on Windows, including the ability to natively load Linux executables and libraries. Canonical will provide bash and all the other command-line tools that are expected in a Linux environment.
Microsoft is positioning WSL strictly as a tool for developers, with a particular view to supporting Web developers and the open source software stacks that they depend on. Many developers are very familiar with the bash shell, with building software using make and gcc, and editing text in vi or emacs. WSL will give these developers versions of these tools that are equal in just about every regard to the ones you get on Linux, because they'll be the ones you get on Linux running unmodified on Windows.
Imaging with CRISPR/Cas9
Deng W, Shi X, Tjian R, Lionnet T, & Singer RH. (2015) CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. Proceedings of the National Academy of Sciences of the United States of America, 112(38), 11870-5. PMID: 26324940
CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells.Nelles DA, Fang MY, O'Connell MR, Xu JL, Markmiller SJ, Doudna JA, & Yeo GW. (2016) Programmable RNA Tracking in Live Cells with CRISPR/Cas9. Cell, 1-9. PMID: 26997482
Programmable RNA Tracking in Live Cells with CRISPR/Cas9.Ubuntu (not Linux) on Windows: How it works
ZDnet: Ubuntu, but not Linux per se, will be running on in the next major Windows 10 update, Redstone.
Overturning bioinformatic predictions, Venter creates minimal synthetic cell with 473 genes
J. Craig Venter and his colleagues have created a synthetic bacterial cell with 473 genes, the number they found are needed for self-replicating life. In doing so, the team has overturned a 20-year-old prediction about how many genes are essential--and revealed major gaps in our knowledge of the roles of these cornerstones of life.
Why open source has been a tremendous accelerator for Monsanto
EnterprisersProject: CIO of Monsanto talks about the two-way win of open source.
Structural Insights into the Processing of Nucleobase-Modified Nucleotides by DNA Polymerases.
Structural Insights into the Processing of Nucleobase-Modified Nucleotides by DNA Polymerases.
Acc Chem Res. 2016 Mar 5;
Authors: Hottin A, Marx A
Abstract
The DNA polymerase-catalyzed incorporation of modified nucleotides is employed in many biological technologies of prime importance, such as next-generation sequencing, nucleic acid-based diagnostics, transcription analysis, and aptamer selection by systematic enrichment of ligands by exponential amplification (SELEX). Recent studies have shown that 2'-deoxynucleoside triphosphates (dNTPs) that are functionalized with modifications at the nucleobase such as dyes, affinity tags, spin and redox labels, or even oligonucleotides are substrates for DNA polymerases, even if modifications of high steric demand are used. The position at which the modification is introduced in the nucleotide has been identified as crucial for retaining substrate activity for DNA polymerases. Modifications are usually attached at the C5 position of pyrimidines and the C7 position of 7-deazapurines. Furthermore, it has been shown that the nature of the modification may impact the efficiency of incorporation of a modified nucleotide into the nascent DNA strand by a DNA polymerase. This Account places functional data obtained in studies of the incorporation of modified nucleotides by DNA polymerases in the context of recently obtained structural data. Crystal structure analysis of a Thermus aquaticus (Taq) DNA polymerase variant (namely, KlenTaq DNA polymerase) in ternary complex with primer-template DNA and several modified nucleotides provided the first structural insights into how nucleobase-modified triphosphates are tolerated. We found that bulky modifications are processed by KlenTaq DNA polymerase as a result of cavities in the protein that enable the modification to extend outside the active site. In addition, we found that the enzyme is able to adapt to different modifications in a flexible manner and adopts different amino acid side-chain conformations at the active site depending on the nature of the nucleotide modification. Different "strategies" (i.e., hydrogen bonding, cation-π interactions) enable the protein to stabilize the respective protein-substrate complex without significantly changing the overall structure of the complex. Interestingly, it was also discovered that a modified nucleotide may be more efficiently processed by KlenTaq DNA polymerase when the 3'-primer terminus is also a modified nucleotide instead of a nonmodified natural one. Indeed, the modifications of two modified nucleotides at adjacent positions can interact with each other (i.e., by π-π interactions) and thereby stabilize the enzyme-substrate complex, resulting in more efficient transformation. Several studies have indicated that archeal DNA polymerases belonging to sequence family B are better suited for the incorporation of nucleobase-modified nucleotides than enzymes from family A. However, significantly less structural data are available for family B DNA polymerases. In order to gain insights into the preference for modified substrates by members of family B, we succeeded in obtaining binary structures of 9°N and KOD DNA polymerases bound to primer-template DNA. We found that the major groove of the archeal family B DNA polymerases is better accessible than in family A DNA polymerases. This might explain the observed superiority of family B DNA polymerases in polymerizing nucleotides that bear bulky modifications located in the major groove, such as modification at C5 of pyrimidines and C7 of 7-deazapurines. Overall, this Account summarizes our recent findings providing structural insight into the mechanism by which modified nucleotides are processed by DNA polymerases. It provides guidelines for the design of modified nucleotides, thus supporting future efforts based on the acceptance of modified nucleotides by DNA polymerases.
PMID: 26947566 [PubMed - as supplied by publisher]
CRISPR Clarifies Split-Hand/Foot

While James R. Clapper, Director of National Intelligence, calls genome editing a “national security threat”, bioethicists warn of CRISPR-created superbabies, and prominent researchers argue whether patents trump papers, I prefer to quietly look at applications of the technology that aren’t dramatic enough to enter the endless news cycle, but elegantly reveal the power of the technology.
I found one such report in the current issue of Genome Research, about ectrodactyly (“absence of digits”), more colorfully known as split-hand/foot malformation. It affects about 1 in 18,000 newborns, and is also seen in toads, frogs, salamanders, mice, rabbits, cows, chickens, marmosets, dogs, cats, and manatees.
LOBSTER PEOPLE
Wikipedia reports cases of “cleft hand” as far back as 1575. Today, a cluster of families with the condition in northern Zimbabwe, who are missing the middle 3 toes and have turned-in outer toes, describe themselves on Facebook as “the ostrich people.”

A more common name is “lobster claw deformity”. The most famous person to compare himself to a crustacean was Grady Stiles Jr, who performed in sideshows as “Lobster Boy.” His father and two of his four children also had it, suggesting autosomal dominant inheritance. American Horror Story: Freak Show used Grady Stiles Jr’s image in the fourth season. The beautiful actress and talk show host Bree Walker refused to keep hiding her hands while reporting the news, and a very recent newspaper article featured pianist Jason Black, aka the Black Scorpion.
Surgery can help, and a gofundme campaign is asking for contributions to treat a baby not yet born.
Although several genes have been implicated in ectrodactyly, the responsible mutation remains a mystery in two-thirds of affected individuals.

In the olden days, before exome sequencing and CRISPR/Cas genome editing, identifying the gene behind a rare single-gene disease relied on finding rare families in whom a detectable chromosomal problem tracked with a specific syndrome. Developing mouse models took years. But using mice and CRISPR/Cas, Stefan Mundlos, MD, of the Max Planck Institute for Molecular Genetics and Guntram Borck, PhD, of the University of Ulm and their colleagues created mice with an array of mutations in a novel gene behind split-hand/foot in under 10 weeks (DNA Science covered Dr. Mundlos’s work on mutations that turn arms into legs.)
A TALE OF TWO FAMILIES
The researchers began with a 6-generation family A, from Pakistan. The analysis hits all aspects of genetics:
- Mendel’s first law
- deducing mode of inheritance from pedigrees bearing the telltale double horizontal lines depicting cousin-cousin couplings
- SNP mapping to narrow down genes to chromosomes
- exome sequencing to nail the responsible mutation
- connecting genes functionally by their expression interactions
A cousin-cousin marriage in generation 4 begat an affected man in generation 5. He married his cousin, leading to an affected girl in generation 6. A different first-cousin pair in generation 5 produced two boys with the condition in generation 6. The pattern of the condition showing up only after consanguineous pairings suggests autosomal recessive inheritance.
Mapping using single nucleotide polymorphisms (SNPs) in family A pointed to a section of the long arm of chromosome 2 not known to harbor any genes associated with limb development. Then exome sequencing of one of the affected boys led to two candidate genes on chromosome 2, only one of which – ZAK – was known to affect limb development, in mice and human embryos. The three cousins each had two copies of a missense mutation in ZAK, which substitutes one amino acid for another in the encoded protein.
Next the researchers sequenced ZAK in 106 unrelated patients, and found a partial deletion in both chromosome 2s of a boy from Tunisia, family B. His parents, who are first cousins, and his brother have only one copy of the deletion and have normal hands and feet. The boy has normal hands but duplicated nail beds, and feet with merged toes (syndactyly) and one duplicated toe. His deletion removes the final 4 of 16 exons in ZAK, and the mutation was not in any databases.
Although family A has a missense mutation and family B a deletion, both affect the same part of the gene, called the SAM domain.

MICE MIMIC THE TUNISIAN BOY
The researchers used mice to further analyze the role of ZAK in split-hand/foot malformation, using the MIT version of CRISPR to make precise deletions, insertions, and swaps.
First, they created mice transgenic for human ZAK. Then they genome edited mouse embryonic stem cells to get animals with one copy of the mutation, and bred them to create the 1 in 4 offspring (Mendel’s first law) that have two copies of the mutation, like the three cousins and the boy from Tunisia.
Mice with both copies of ZAK entirely knocked out didn’t survive beyond the embryo stage. So people with split-hand/foot due to mutations in ZAK must have less catastrophic mutations than the doomed mice, because they survive to be born.
What would happen if the mice only had problems with the SAM parts of their ZAK genes, like the people? Sure enough, mice with SAM edited out had doubled or stunted hindlimbs or feet, split femurs, and extra toes. Ectrodactyly is also variable in humans, even within families like that of the original Lobster Boy.
Next, the researchers probed the genes with which ZAK interacts. They found very little expression of a gene called Trp63, known to cause ectrodactyly in people, in the mice with the weird legs.
Remember the Pakistani cousins? Modeling the ZAK protein (using a technique called circular dichroism) showed that the family’s single-base mutation unravels alpha helices that are normally nestled into the core of the SAM domain away from water. That tiny glitch destabilizes SAM and ZAK. Rather than neatly self-assembling, they glom together in a way that prevents development of normal limb “buds”.
Many details remain to be worked out, but this paper is an important step in unraveling gene interactions that oversee limb formation.
Alas, we have a long history of scary biotechnologies becoming commonplace over time. Perhaps CRISPR/Cas and other genome editing tools are scarier because they can be deployed on a human germline, given some tricky manipulations. But the bigger picture from the study: genome editing can recapitulate human genetic disease in an animal model in mere weeks.
The researchers conclude that the approach “holds the promise to contribute to a paradigmatic shift in the investigation of Mendelian disorders. This will be extremely useful in the future to investigate the pathogenicity of rare missense alleles that might occur only in single families.”
I wish the media would pay as much attention to these exciting new reports on the fledgling eclectic uses of genome editing as they do the potential misuses.
Oxford Nanopore Updates: Reveals the nanopore used in Nanopore devices
Storified tweets from Oxford Nanopore’s technology focus Google hangout by Clive Brown. Briefly,
- Oxford Nanopore revealed the pore it uses in its devices. It is not MSP pore. :) It is using CsgG pore from E.coli. And it is using heavily engineered CsgG with over 700 mutations (with license) to make it a better pore for sequencing. Immediately after the update meeting, VIB/Vrije Universiteit Brussels announced that VIB’s Han Remaut has teamed up with Oxford Nanopore Technologies for the development of nanopore sensing technology. Han Remaut group has been working on work on the protein nanopore, CsgG and will be in ‘R9’ in the MinION handheld DNA sequencer and the PromethIONTM system.
- Oxford nanopore is seeing a big improvement in accuracy with R9 and new base caller. 85% accuracy in 1D and 95% accuracy in 2D. And Oxford nanopore is moving away from the current HMM-based base caller to Neural Network based base caller.

- March-April is pretty hectic for Nanopore, with lots of releases from Oxford Nanopore including the release of source code for base calling, Developmental R9, and first shipping of Promethion device.
Watch the Google hangout recording at You Tube.
Illumina Sues Oxford Nanopore
In case you missed it, Illumina announced that it is Oxford Nanopore Technologies for violating Illumina’s two patents on nanopore sequencing in Oxford Nanopore’s MinION and PromethION devices.
Illumina press release says that
The lawsuits are based on U.S. Patent Nos. 8,673,550 and 9,170,230, which are entitled “MSP NANOPORES AND RELATED METHODS.”Illumina has exclusively licensed the patents in the field of nucleic acid sequencing from the UAB Research Foundation and the University of Washington. The lawsuits focus on ONT’s MinION and PromethION devices.
Illumina has made substantial investments to obtain licenses and develop the nanopore sequencing technology invented by researchers at the University of Alabama at Birmingham and University of Washington. Illumina filed the lawsuits to protect its investment and patent rights in this technology.
Swiftly, Oxford Nanopore shot back in a press release, where Dr Gordon Sanghera, CEO of Oxford Nanopore said
It is gratifying to have the commercial relevance of Oxford Nanopore products so publicly acknowledged by the market monopolist for NGS.
We do not anticipate any disruption to our ongoing commercial progress as a result of Illumina’s action, which we believe is without merit.
The two patents in question here is on the molecule Mycobacterium smegmatis Porin (MSP), that Oxford Nanopore could be using as the nanopore for sequencing. M. smegmatis Porin is a beautiful protein made of beta sheets.
The two patents named “MSP nanopores and related methods”, were filed by from UW Washington and Univ. Alabama Birmingham.
Oxford Nanopore has never mentioned what types of nanopores is used in MinION and PromethION devices. However, early on, the founders of Oxford Nanopore has mainly used alpha Hemolysin as a pore for sequencing. One of their early demonstrations of Nanopore DNA sequencing in 2009 used alpha Hemolysin mutant form.
- Continuous base identification for single-molecule nanopore DNA sequencingNature Nanotechnology DOI: 10.1038/nnano.2009.12, James Clarke, Hai-Chen Wu, Lakmal Jayasinghe, Alpesh Patel, Stuart Reid, Hagan Bayley (2009)
Oxford Nanopore also seems to have patents on other types of pores including graphene based solid-state nanopores and mutant Mspa. One of the Oxford Nanopore’s patents, titled “Mutant pores“, is all about mutant forms of Mspa. The patent seems to indicate that Hemolysin pores show high variance in observed current.
While the current range for nucleotide discrimination has been improved through mutation of the hemolysin pore, a sequencing system would have higher performance if the current differences between nucleotides could be improved further. In addition, it has been observed that when the nucleic acids are moved through a pore, some current states show high variance. It has also been shown that some mutant hemolysin pores exhibit higher variance than others. While the variance of these states may contain sequence specific information, it is desirable to produce pores that have low variance to simplify the system. It is also desirable to reduce the number of nucleotides that contribute to the observed current.
The patent goes to claim that multiple mutations on Msp porins seem to “strikingly” improve nucleotide discrimination, low variance, and high signal-to-noise ratio.
The inventors have surprisingly demonstrated that novel mutants of Msp display improved properties for estimating the characteristics, such as the sequence of nucleic acids. The mutants surprisingly display improved nucleotide discrimination. In particular, the mutants surprisingly display an increased current range, which makes it easier to discriminate between different nucleotides, and a reduced variance of states, which increases the signal-to-noise ratio. In addition, the number of nucleotides contributing to the current as the nucleic acid moves through the pore is decreased. This makes it easier to identify a direct relationship between the observed current as the nucleic acid moves through the pore and the nucleic acid sequence. The inventors have also surprisingly shown that Msp shows improved sequencing properties when the movement of the nucleic acid through the pore is controlled by a Phi29 DNA polymerase.
It is interesting see that the 2012 Nature Biotechnology paper using MSP pore for sequencing by Manrao et al from Jens H Gundlach group acknowledges M. Akeson and G.M. Cherf for their help with phi29 DNA-polymerase.
We thank M. Akeson and G.M. Cherf for getting us started with the blocking oligomer phi29 DNAP technique, sharing their CAT DNA and reading the manuscript.
The same Nature Biotechnology issue also has the paper from M. Akeson group demonstrating sequencing with alpha-Hemolysin pore and around the same time Oxford Nanopore made the big announcement on their products at AGBT.
It is really sad to see such a lawsuit, especially the idea of nanopore sequencing has been around for a while and just when Oxford Nanopore seems to gain a lot of traction in the last few year. Here are links to some other interesting blog posts on the lawsuit and comments on Twitter.
- Mick Watson: Shots fired – nanopore wars part II
- BioIT World: Illumina Sues Oxford Nanopore Technologies Over Composition of Nanopores
Sad @illumina employing patent litigation as a weapon against competitor @nanopore; they did same against @CompleteGenomic during their IPO
— Daniel MacArthur (@dgmacarthur) February 25, 2016
First they laugh at you, then they sue you:@nanopore https://t.co/ekvNyKqeiS
— Yaniv Erlich (@erlichya) February 24, 2016
Generation of a pair of independently binding DNA aptamers in a single round of selection using proximity ligation.
 |
Related Articles |
Generation of a pair of independently binding DNA aptamers in a single round of selection using proximity ligation.
Chem Commun (Camb). 2015 May 28;51(43):9050-3
Authors: Chumphukam O, Le TT, Piletsky S, Cass AE
Abstract
The ability to rapidly generate a pair of aptamers that bind independently to a protein target would greatly extend their use as reagents for two site ('sandwich') assays. We describe here a method to achieve this through proximity ligation. Using lysozyme as a target we demonstrate that under optimal conditions such a pair of aptamers, with nanomolar affinities, can be generated in a single round.
PMID: 25941004 [PubMed - indexed for MEDLINE]



